The uncoupling of catalysis and translocation in the viral RNA-dependent RNA polymerase
Date:05-04-2017 | 【Print】 【close】
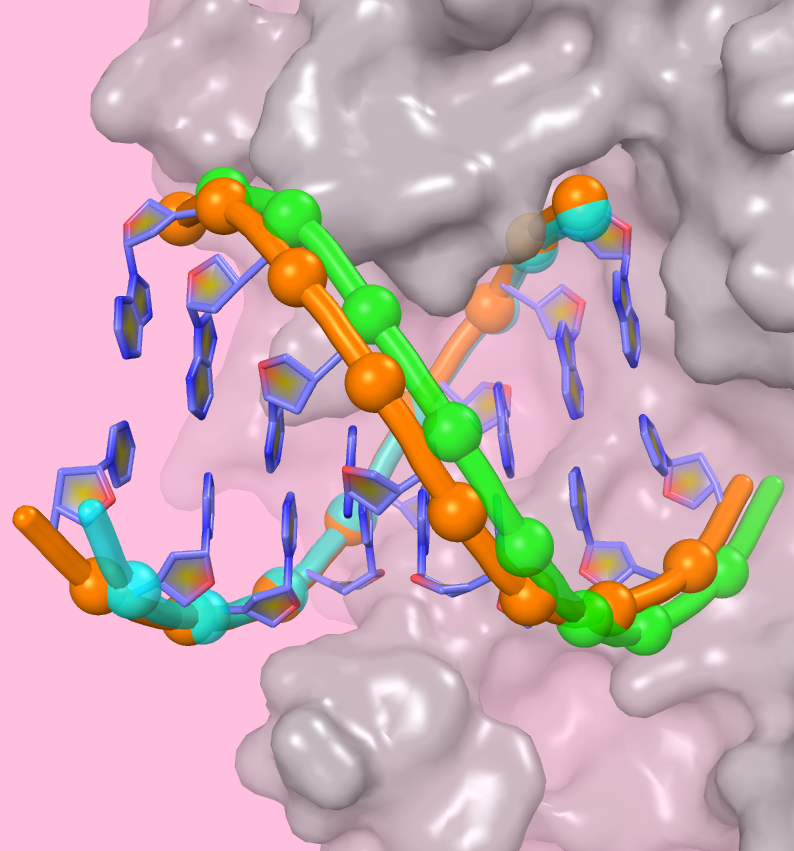
The nucleotide addition cycle of nucleic acid polymerases includes two major events: the pre-chemistry active site closure leading to the addition of one nucleotide to the product chain; the post-chemistry translocation step moving the polymerase active site one position downstream on its template. In viral RNA-dependent RNA polymerases (RdRPs), structural and biochemical evidences suggest that these two events are not tightly coupled, unlike the situation observed in A-family polymerases such as the bacteriophage T7 RNA polymerase.
Recently, in a paper written by Prof. Peng Gong and Dr. Shu Bo in WIV and published on RNA Biol., an RdRP translocation intermediate crystal structure of enterovirus 71 shed light on how translocation may be controlled by elements within RdRP catalytic motifs, and a series of poliovirus apo RdRP crystal structures explicitly suggest that a motif B loop may assist the movement of the template strand in late stages of transcription. Implications of RdRP catalysis-translocation uncoupling and the remaining challenges to further elucidate RdRP translocation mechanism are also discussed.
Contact:
GONG Peng
E-mail: gongpeng@wh.iov.cn
Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China 430071(http://english.whiov.cas.cn/)